この記事は日本語の記事があります(This article is available in Japanese)
MAFFT→RAxML→FigTreeで遺伝子の系統樹を作成する – Ubuntu19.04
Problem
After analyzing with RAxML, I wanted to make phylogenetic tree with “FigTree” software, and I made it but I could’t make phylogenetic tree with bootstrap values.
Solution
RAxML operation
First of all, you should enter below commands. (If you use raxmlHPC-PTHREADS-AVX and analyzing amino acid sequences. You should change that part of command according to your environment.)
raxmlHPC-PTHREADS-AVX -f b -m PROTGAMMAAUTO -t RAxML_bestTree.HOGE -z RAxML_bootstrap.HOGE -n BOOTSTRAP“-f b” option means “drawing bipartition information on a tree”
“-m” means “model”. In this command, I used “AUTO” model for amino acid sequences, so you should choose best model for your data.
“-t” option requires “tree data file”. You may choose “RAxML_bestTree” as ML tree.
“-z” option requires “bootstrap data”. You may choose “RAxML_bootstrap”.
“-n” option means identifier, as you did before.
After enter and execute it, you can find new 3 files in the directory with “.BOOTSTRAP”, and “RAxML_bipartitions.BOOTSTRAP” is object.
How to make phylogenetic tree with FigTree?
You can get FigTree software here.
After execute the application and open “RAxML_bipartitions.BOOTSTRAP” , a dialog will be opened like below.
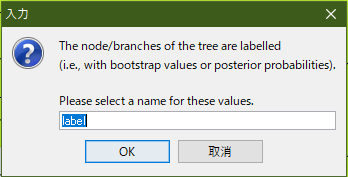
You should name “Bootstrap”, and click “OK”.
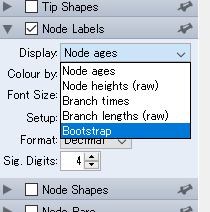
After loading a phylogenetic tree, see the left side bar, and you can find “Node Labels”. Click the tab, and click the drop-down list and choose “Bootstrap”.
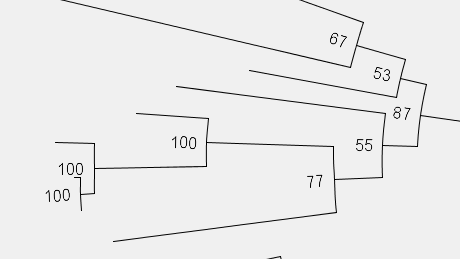
Done!


![【Qiskit】マルチオミクス解析を量子機械学習でやる①[環境構築・基礎]](https://kimbio.info/wp-content/uploads/2024/05/2203027-100x100.jpg)



ピンバック:MAFFT→RAxML→FigTreeで遺伝子の系統樹を作成する - Ubuntu19.04 - Kim Biology & Informatics